Benchmarking Mojo, Julia, and Python + NumPy
Published:
 Image: the Julia set for $f(z) = z^4 + 0.6 + 0.55i$, which the author could not elegantly work into this post.
Image: the Julia set for $f(z) = z^4 + 0.6 + 0.55i$, which the author could not elegantly work into this post.
The freshly-announced Mojo programming language has made some ambitious claims about its performance. The language’s creators have suggested that it can be up to 35,000x faster than Python. This eye-popping statement can be retethered to reality by observing that this speed increase compares fully optimized Mojo to vanilla Python. Since most Python computations are done with packages written in C/C++, practical improvement should be smaller.
Nevertheless, the speed increase is significant enough that it merits further investigation. Some community members have already taken up this task and compared Mojo to Python code written with NumPy or numba, two of the more popular performance-oriented Python packages. I’d like to take this a step further and compare Mojo to its natural enemy: Julia.
What we are unable to do, currently, is run Mojo and Julia on the same hardware. As of writing, Mojo is not publicly available and can only be run in the Mojo playground web environment. I can, however, run Python both in the playground and on my personal machine. Therefore we can compare Mojo and Julia to Python and ask which gives more lift.
To this end, we will implement the Mandelbrot set in all three languages and compare their execution times.
Results
Here’s what you came for.
Local computations
| Language | Execution time (ms) | Improvement over Python |
|---|---|---|
| Python (numpy) | 1997.51 | 1x |
| Julia unoptimized | 141.23 | 14.1x |
| Julia paralellized | 93.52 | 21.3x |
| Julia optimized | 33.65 | 59.4x |
Mojo notebook computations
| Language | Execution time (ms) | Improvement over Python |
|---|---|---|
| Python (numpy) | 735.7 | 1x |
| Mojo SIMD | 10.36 | 71x |
| Mojo optimized | 4.76 | 154.5x |
Since the Mojo notebook ran on 32 threads and my Julia code ran on 8, it’s hard to say with confidence that the Mojo implementation is 3x faster than Julia. It may not be faster at all!
Dubious experimental methodology notwithstanding, it’s clear that Mojo has a lot to offer in terms of performance. The downside, at present, is that it’s complicated to attain those performance benefits. Julia, in comparison, makes it absurdly easy to boost performance. All I had to do to improve very basic Julia code was add three macros, one of which was from the Julia base.
I predict that the Julia vs. Mojo war will be conducted on the battlefield of simplicity. If Mojo is able to provide high-level abstractions to easily leverage techniques like SIMD and parallelization, then Mojo will be a serious contender for scientific computing supremacy. Since providing these abstractions seems to be, more-or-less, an explicit goal of Mojo, I think its outlook is bright.
Adoption is king, and Julia fans are (rightly) worried that Mojo will leech away some of its potential userbase. There is still room for both, though: scientific computing is not a stated target domain for Mojo; its focus is AI infrastructure. The scientific computing community is notoriously resistant to change (many people still use MATLAB, of all things) and the benefits of Mojo, in terms of pure scientific computing, may prove too negligible to be a significant draw.
The NumPy baseline
After some experimention, this is the fastest I could do in NumPy:
import numpy as np
def mandelbrot_kernel(c, maxiter):
z = np.copy(c)
iter_count = np.zeros(c.shape, dtype="int")
for i in range(maxiter):
mask = np.abs(z) < 2
if not np.any(mask):
break
z[mask] = z[mask]**2 + c[mask]
iter_count[mask] = i
return iter_count
def compute_mandelbrot(xmin, xmax, ymin, ymax, width, height, maxiter):
x = np.linspace(xmin, xmax, width)
y = np.linspace(ymin, ymax, height)
c = x[np.newaxis, :] + y[:, np.newaxis] * 1j
return mandelbrot_kernel(c, maxiter)
The only pure Python loop occurs in the mandelbrot_kernel as we repeatedly apply the function $z \mapsto z^2 + c$.
I measured execution time using time.perf_counter_ns():
t0 = time.perf_counter_ns()
q = compute_mandelbrot(-2.0, 0.6, -1.5, 1.5, 960, 768, 200)
t1 = time.perf_counter_ns()
On my machine:
>>> print(f"{(t1-t0)/1e6} ms")
1997.505474 ms
In the Mojo playground, executing Python is dead-simple: just add %%python at the top of a cell, and it executes as Python rather than Mojo. Note: I have no idea how the Mojo kernel runs Python, so there could be improvements/slowdowns in this process.
In the Mojo playground:
>>> print(f"{(t1-t0)/1e6} ms")
735.645235 ms
So the Mojo playground is roughly 3x faster than my machine.
Julia
It was quite easy to beat Python using Julia. Just some plain Julia with no optimizations ran in 260ms on my machine, or nearly 8x faster than the NumPy implementation.
Adding some loop vectorization and parallel computing macros was both easy and effective. Here’s the final code.
using BenchmarkTools
using Base.Threads
using LoopVectorization
function mandelbrot_kernel(c::ComplexF64, maxiter::Int64)
z = 0
@inbounds @fastmath for i in 1:maxiter
if abs(z) > 2
return i-1
end
z = z^2 + c
end
return maxiter
end
function compute_mandelbrot(xmin::Float64, xmax::Float64, ymin::Float64, ymax::Float64, width::Int64, height::Int64, maxiter::Int64)
x = LinRange(xmin, xmax, width)
y = LinRange(ymin, ymax, height)
k = Matrix{Int}(undef, height, width)
@threads for i in 1:width
for j in 1:height
k[j, i] = mandelbrot_kernel(complex(x[i], y[j]), maxiter)
end
end
return k
end
I then timed it with BenchmarkTools.jl:
>>> @btime compute_mandelbrot(-2.0, 0.6, -1.5, 1.5, 960, 768, 200)
33.653 ms (50 allocations: 5.63 MiB)
This is run with 8 threads on my machine and whatever pitiful SIMD is available. I’ll note that parallelizing cut the time roughly in half, @inbounds did almost nothing, and @fastmath did the rest.
Fastmath is a little sketchy. From the docs:
@fastmath expr
Execute a transformed version of the expression, which calls functions that
may violate strict IEEE semantics. This allows the fastest possible operation,
but results are undefined -- be careful when doing this, as it may change numerical
results.
...
The program ran in 93.52 ms without fastmath, which may be a better number to compare with.
Plotting the set is very easy with Plots.jl.
using Plots
mandyB = compute_mandelbrot(-2.0, 0.6, -1.5, 1.5, 960, 768, 200)
heatmap(mandyB, aspect_ratio=:equal, color=:grays, xticks=[], yticks=[], cbar=false, dpi=200)
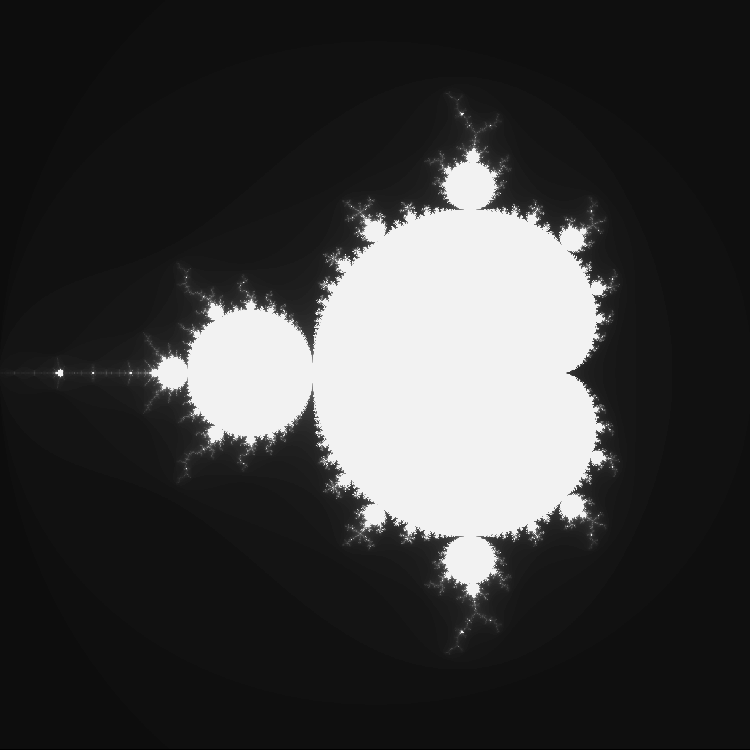
Hello, Mojo
For Mojo, I copied the code directly from the Mandelbrot notebook, which you can browse for yourself. It takes quite a bit more work than the NumPy and Julia implementations.
Interestingly, the notebook ran on 32 threads, rather than the 8 that the demo notebook had. I guess they’ve souped up the hardware since writing their article.
Anyway, the fully vectorized and parallelized Mojo notebook ran in 4.76 ms. The unparallelized (but SIMD-optimized) version ran in 10.36 ms.
